Yes, i searched in the raw data

Personalized Nutrition
#31
Posted 06 December 2015 - 07:49 PM
#33
Posted 07 December 2015 - 05:46 PM
Nice interview, via The Quantified Body link, with Dr. Michael Nova, Chief Innovation Officer and Founding Executive at Pathway Genomics, offering genetic testing services from screening to precision health. Their partnership with IBM can possibly lead to a train Watson AI for personalized nutrition as foreseen in one of my previous post here.
https://thequantifie...h-michael-nova/
- Dr. Nova’s roots in genetics and how Pathway Genomics differs from 23andMe in structure, technology, staff, and interpreting testing results (06:12).
- Why reporting on genetic tests varies between companies; why testing does not produce ‘black and white’ interpretations of tested parameters (15:22).
- The meaning of personalized / precision medicine; current applicability and future prospects, as numerous testing technologies become cheaper (17:46).
- How genetic test panels are researched and converted into actionable information for physicians and individuals (20:40).
- The complexity of genetic and epigenetic tests and why professional guidance is required when making health decisions based on results (29:30).
- Why epigenetics is more complex than genetics and how genes are switched on / off by interactions with the environment or due to behavior (33:50).
- Pathway Genomics and IBM’s Watson collaboration – integrating extremely diverse and data-dense medical information into meaningful outputs (36:11).
- How genetic testing improves pharmacological prescription decisions and why increasingly complex data is even more useful (39:20).
- Optimizing exercise for individuals using genetic information (46:04).
- How to access information about personalized medicine and genetic testing (47:33).
- What information Dr. Nova tracks on himself and why it is crucial to be aware of your genetics (49:46).
#34
Posted 08 December 2015 - 03:22 PM
It looks like the early versions of the chip included this SNP but the later v3 /v4 do not.
I have done my 23andMe test in late 2013 and unfortunately it is not included.
#35
Posted 08 December 2015 - 05:02 PM
It looks like the early versions of the chip included this SNP but the later v3 /v4 do not.
I have done my 23andMe test in late 2013 and unfortunately it is not included.
Yes, it must be so. I have a somewhat reverse problem with my test done as early as 2011. E.g. I was looking at rs603424 which is "not genotyped" for me but actually other folks have it reported. It is an important SNP in the SDC1 gene strongly influencing the free fatty acids palmitic to palmitoleic ratio I wish to research on for me. I did not do it yet but some recommend to use DNA.LAND to infer it. I posted before on this and if I am successful in getting information on this SNP I will let you know how the process works. You might wish to try for yourself.
#36
Posted 08 December 2015 - 06:55 PM
23andme say that this is a SNP for the PKD2L1 Gene (not SDC1) ![]()
#37
Posted 08 December 2015 - 10:02 PM
23andme say that this is a SNP for the PKD2L1 Gene (not SDC1)
Sorry, I misspelled in my previous post: I meant SCD1 (not SDC1). Here is the reference I am considering (see Table 1 and Figure 1, I attach the parts on SCD):
Human metabolic individuality in biomedical and pharmaceutical research
http://www.ncbi.nlm....les/PMC3832838/
 SCD.PNG 267.35KB
1 downloads
SCD.PNG 267.35KB
1 downloads
 rs603424.PNG 97.42KB
1 downloads
rs603424.PNG 97.42KB
1 downloads
I understand that rs603424 in SCD1 influences the palmitic / palmitoleic fatty acid ratio (C16:0 / C16:1 n7) which I am interested in. Not sure what is the link with PKD2L1, I will check.
Edited by albedo, 08 December 2015 - 10:05 PM.
#38
Posted 13 December 2015 - 03:46 PM
Aurel, Aribadabar, Sthira,
Just wish to let you know I tested the process of DNA.LAND which turned out simple and clear to apply. Again, it allows by statistical inference to deduce a unknown genotype, e.g. the one we could not find in our 23andMe report ("not-genotyped"). The process is called as genetics imputation. E.g. my rs603424 turned out to be GA (mutated) with high likelihood. Allow one day to go through the full (registration) process which has to manage large files. It is a free service and as counter part you will help research. You might google for more information, e.g. see http://www.nature.co...na-land-1.18514
#39
Posted 13 December 2015 - 04:35 PM
Aurel, Aribadabar, Sthira,
Just wish to let you know I tested the process of DNA.LAND which turned out simple and clear to apply. Again, it allows by statistical inference to deduce a unknown genotype, e.g. the one we could not find in our 23andMe report ("not-genotyped"). The process is called as genetics imputation. E.g. my rs603424 turned out to be GA (mutated) with high likelihood. Allow one day to go through the full (registration) process which has to manage large files. It is a free service and as counter part you will help research. You might google for more information, e.g. see http://www.nature.co...na-land-1.18514
Hey Albedo,
as per your suggestion I also uploaded my 23andMe genome file there and they processed it and generated a . vcf and a.bcf file.
How can I find out what the "non-genotyped" SNPs are from these files ( they are 400MB+ each)?
Thanks!
Edited by aribadabar, 13 December 2015 - 04:36 PM.
#40
Posted 13 December 2015 - 06:31 PM
Aribadabar, the process is described in the link "learn more" next to the downloaded vcf file. You basically need to open the file (and search your rs) using the "glogg" program (you have the installation link in the description of the process which is given for Mac or PC). Here is the image I get when I search in the file rs603424. The "learn more" link also has a section "understanding VCF value" with the interpretation of the symbols. To keep on topic please feel free to PM me if you still have issues.
 dnaland.PNG 3.64KB
1 downloads
dnaland.PNG 3.64KB
1 downloads
 rs603424.PNG 4.07KB
0 downloads
rs603424.PNG 4.07KB
0 downloads
#41
Posted 16 December 2015 - 09:46 PM
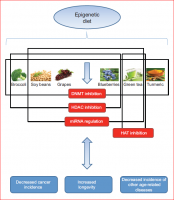
A recent review on the linkage between nutrition, cancer and aging. Possibly, studying the mutations we bring on the many genes mentioned in the article can shed light on how we are individually aging and potentially can point to interventions allowing us to catch the SENS train on time....
" ... Consumption of foods that modulate epigenetic mechanisms has been shown to decrease the incidence of cancer and increase longevity, as well as prevent the onset of other age-related diseases. Cruciferous vegetables, such as broccoli, which are rich in sulforaphane, can act as HDAC inhibitors, regulating certain
cancer-related genes. Genistein, which is found in soy beans, also exhibits chemopreventive properties and can result in both the partial reversal of aberrant DNA
hypermethylation and the regulation of key miRNAs. Grapes contain resveratrol, a phenol that activates SIRT1 (a known HDAC inhibitor) and increases longevity, mimicking the effects of caloric restriction..." (Fig. 3)
http://jeb.biologist...8/1/59.abstract
#42
Posted 17 December 2015 - 03:55 PM
Personalized Nutrition by Prediction of Glycemic Responses
 Personalized Nutrition.PNG 150.27KB
4 downloads
Personalized Nutrition.PNG 150.27KB
4 downloads
"People eating identical meals present high variability in post-meal blood glucose response. Personalized diets created with the help of an accurate predictor of blood glucose response that integrates parameters such as dietary habits, physical activity, and gut microbiota may successfully lower postmeal blood glucose and its long-term metabolic consequences."
http://www.cell.com/...8674(15)01481-6
#43
Posted 17 December 2015 - 07:43 PM
Albedo, have you - beside the 23andme test - taken any blood tests of certain parameters? If yes, then which ones and why? Thanks!
#44
Posted 18 December 2015 - 12:59 PM
Albedo, have you - beside the 23andme test - taken any blood tests of certain parameters? If yes, then which ones and why? Thanks!
Yes Aurel, I have blood test every year and for some markers more frequently depending on the risks and/or trying to assess if what I do works. Please challenge and of course take this only for educational/research purposes.
They are just too many to list but I feel I should mention, with particular focus on nutritional pro-action:
- Haematology Profile (aka Complete Blood Count). I look for any abnormality. E.g. I tend to have a lower no of leucocytes (I had leucopenia disease in my family) and it can indicates some copper deficiency (I also supplement with zinc). I had it (normally) higher during infection, post-surgery etc… I used once a course of shark liver oil and probably have seen something but discontinued.
- Haemostatics. I look in particular at Fibrinogen. I tended to have it higher (more clotting and risker) but normalized when I decided to take better care of myself with lifestyle and nutrition changes.
- Clinical chemistry. Checking for cardiovascular risks, metabolic function, electrolyte status, minerals, liver and kidney function. I check in particular my iron status. I had large values of Ferritin so that I wondered about genetic haemochromatosis but I am not a carrier (check in particular rs1800562 where I am GG). I avoid iron and supplement with IP6 (inositol hexaphosphate) which looks for me a good iron chelator while phosphorus is in range. I am back to normal for Ferritin. I am still trying to correct my red blood cell (RBC) magnesium which is abnormally low. I test for glucose, HbA1C and Homeostasis Model Assessment for insulin sensitivity (HOMA). I tend to be a bit high for HbA1C and I wish to see an effect from my anti-glycation supplementation (e.g. carnosine, not seen so far) and carrying a test with low dose 250mg metformin for other reasons (anti-aging). I look in particular to total Cholesterol, LDL, HDL, ApoA/ApoB ratio, Lp(a), Triglycerides (TG) and passed two times a LDL-cholesterol density patterns and particle size (VAP) test. I have (primary vs. familial) dislipedia hence actionable with lifestyle and nutrition. I watch for dietary cholesterol and supplement with red yeast rice which contains naturally a statin. I have it hard but the good news is my lifestyle actions changed the LDL size pattern from the atherosclerotic risky pattern B (small, dense LDL) to the less risky pattern A (large, buoyant LDL). My relatively low carbs diet change strongly reduced TG. I could never get homocysteine less than 10 (but had it at 16!, most likely due to supplementing with niacin, removed since) and will make a test using 1000mcg of l-methylfolate. I look at (chronic) inflammation, keeping hr-CRP as low as possible (0.1-0.4). I try to keep (easy) my Vitamin D 25 Hydroxy around 40 ng/ml (I do not like much higher than that as some proposes (e.g. > 50), there are links to atrial fibrillation which is one of my genetic risks).
- Vitamin status. From time to time I include some B group tests to check my supplementation (e.g. B12, as I use metformin which might reduce it but more likely at higher doses and I genetically also have a reduced odd of response which might make me increase the dose, and folates) and cross-check with my homocysteine level.
- Free fatty acids. I look in particular at the pro-inflammatory arachidonic acid (AA), EPA, DHA, several ratios (e.g. EPA/AA indicative of inflammation, palmitoleic /palmitic which I already mentioned in this thread) and the omega-3 index.
- Hormonal. I have a prostate condition and increased risk of cancer. I monitor my free-testosterone for the obvious reasons and in particular DHT for which a large value is a risk factor for enlarged prostate. I look at thyroid (for metabolism and I use soy foods which is risky for the gland), insulin and IGF-1. I tend to test very low for DHEA but avoid, following many tries, supplementation.
- Cancer. I test regularly some tumor markers, in particular total and free PSA (where I watch mostly the slope in time, aka velocity) for my prostate condition and others such as CEA and CA. I tested (in the US) a couple of times for AMAS (Anti-Malignan Antibody) an early detector. I also included a couple of times the HCG-human chorionic gonadotrophin (Dr Xandria Williams in “Vital Signs for Cancer Prevention” makes a good case for it).
I hope this helps. I am surely forgetting a lot more; please feel free to challenge this and include what should be tested as an actionable (e.g. via nutrition) biomarker.
#45
Posted 18 December 2015 - 04:02 PM
Thank you for taking the time to write this. I follow your thread with great interest. I asked because recently I had a 23andme test and start reading into the evaluation of the different services, especially at the non-common abnormalities with high impact on health. One for example was, that some cycle could be working differently thus producing more ammoniac in the body. So I got this tested with some other markers. Like you I think it is good to have a history of blood test, thus hopefully one can find out what is normal for yourself (and not rely on the reference frames).
Very interesting. I do hope that our health system will jump on board with this, as preanalytics will be much cheaper than treating an already ill person.
#46
Posted 19 December 2015 - 01:49 PM
I keep looking at the news these days on Nicotinamide Riboside (NR). Probably the best place here to learn about it is Bryan_S’s thread (curated):
http://www.longecity...boside-curated/
Bryan_S pointed to an interesting and very recent paper:
“…A decrease in intracellular levels of nicotinamide adenine dinucleotide (NAD+) has been shown to occur during the aging process. This decrease in NAD+ levels has a causal role on the development of age-related mitochondrial dysfunction and metabolic decline. To date, the mechanisms responsible for the age-related NAD+ decline have not been identified. It has been proposed that accumulation of DNA damage driven PARP activation may be involved. However, we identify that PARP levels and activity decline with aging. In contrast, we demonstrate for the first time that the expression and activity of the enzyme CD38 increases with aging and plays an active role in the age-related NAD+ decline in vivo and the subsequent development of age related mitochondrial dysfunction. In addition, we also identify CD38 as the main enzyme involved in the degradation of NAD+ precursors such as nicotinamide mononucleotide (NMN) in vivo and to have a role in the modulation of the response to NAD+ replacement therapy in aging. These data demonstrates the key role of CD38 in age-related NAD+ and metabolic decline, and highlights the potential role of CD38 inhibition for the development of an effective “NAD+ replacement therapy" for aging and other metabolic diseases…”
I found that post very informative also because there are studies looking at genotype profiles in healthy humans which can tell about the CD38 activity and possibly point to therapeutic interventions. E.g.:
“ .. CD38 (EC 3.2.2.6, NAD(+)-glycohydrolase) is a multifunctional enzyme catalyzing the synthesis and hydrolysis of cyclic ADP-ribose from NAD+ to ADP-ribose. The loss of CD38 function is associated with impaired immune responses, metabolic disturbances, and behavioral modifications. Notably, it has been linked to HIV infection, leukemias, myelomas, solid tumors, Type II Diabetes mellitus, bone metabolism, as well as Autism Spectrum Disorder. Taking into account the crucial role played by CD38 in many diseases and in clinical practice, here we assessed the distribution of CD38 NADase activity in a healthy population (104 sex-matched unrelated individuals, 12–98 years) and determined its main predictors among genetic and physiological factors (age and sex). The mean value of CD38 NADase activity was 0.051±0.023 mU/mg (0.010–0.099 mU/mg), following a normal distribution in the study population (Kolmogorov–Smirnov test P=0.200). The TNF-α -308G>A (rs1800629) resulted the main predictor (β=0.364, P=0.00008), followed by Age (β=0.280, P=0.002) and the CD38 184C>G (rs6449182) (β=0.193, P=0.033). Our study contributes to understanding CD38 enzyme physiological functions, by reporting, for the first time, its activity distribution in healthy individuals and demonstrating a significant positive correlation with age. Moreover, the possible use of TNF-α -308G>A (rs1800629) and the CD38 184C>G (rs6449182) SNPs as predictive genetic markers of CD38 activity, clearly point toward possible pharmacogenomic applications and to a more refined use of CD38 in clinical settings…”
I could get hold of the previous paper and tried to look at my genotype for the SNPs which are indicated therein, trying to answer, even if only qualitatively though, the question whether or not I could benefit from NR.
I assume understanding correctly that higher CD38 activity would mean decreased intracellular NAD (as confirmed in the paper by CD38-/- mouse having 10-20x higher NAD). CD38 also looks reducing SIRT1.
Looking at the Table 2 (attached) with the best predictors of CD38 activity, I found out that being GG for TNF-α -308G>A (rs1800629) which is the 1st best predictor (p=0.00008) tells me basically nothing useful to reply my question.
 CD38.PNG 56.58KB
4 downloads
CD38.PNG 56.58KB
4 downloads
However, my age (2nd best predictor) and being CG for CD38 184 C>G (rs6449182) (3rd best predictor), therefore bringing the G allele which increases the CD38 expression, would presumably imply together a benefit with NR supplementation. Moreover, not bringing the rs1800561 SNP (I am CC) for which the paper reports a 50% decrease in CD38 activity, would add to the argumentation.
Of course, this is qualitative as the age and the effect of the second part of the genotype (the C allele in rs6449182) should be disentangled and quantitatively assessed. It might also be that, again qualitatively, the C allele of rs6449182 would make high doses of NR are not required as the activity of CD38 is already partially reduced. Just for the record I am running today at 200 mg of NR (with 250 mg t-resveratrol and 25 mg of pterostilbene).
To me this looks like another good example of a “personalized nutrition” intervention, here with NR.
#47
Posted 19 December 2015 - 04:27 PM
The paper is available free here: https://www.research...CD38_184CG_SNPs
#48
Posted 23 December 2015 - 01:40 PM
....
(2) Carriers of two copies of rs7946 variants V175M on PEMT are also about 2x more likely to have NAFLD than non carriers. E.g. see:
Polymorphism of the PEMT gene and susceptibility to nonalcoholic fatty liver disease (NAFLD).
I am TT and so I might be more susceptible to NAFLD which is probably the case with my brother?
A 3rd studied variant, rs12325817 on PEMT, related to estrogen response, is unfortunately not reported by 23andMe and I am looking how I can infer that information too to support my point.
The DNA.LAND imputation file provides rs12325817 as CC (ref/ref) with the alt allele being G. So I look not carrying that mutation. The study of rs12325817 is from S. Zeisel:
Nutritional Genomics: Defining the Dietary Requirement and Effects of Choline1,2,3
http://www.ncbi.nlm....les/PMC3040911/
"...Most men and postmenopausal women, but only 44% of premenopausal women, fed low-choline diets developed reversible fatty liver (measured by mass resonance spectroscopy) as well as liver and muscle damage (16). This difference in dietary choline requirement for young women occurs, because they have an estrogen-enhanced capacity for producing their own choline; the PEMT gene (forms phosphatidylcholine) is induced by estrogen in human hepatocytes, with maximal activation at estrogen concentrations reached at term in pregnancy (17). Thus, capacity for this endogenous source of choline is highest during the period when females need to support fetal development. This is important, because the demand for choline is very high during pregnancy and lactation (18) and, as discussed later, choline is critical for normal fetal development.Though more than one-half of premenopausal women were resistant to choline deficiency-induced organ dysfunction, those premenopausal women that did require a dietary source of choline had a SNP in PEMT (rs12325817; 74% of women in North Carolina had 1 or more variant alleles), making them unresponsive to estrogen induction of PEMT (19, 20). This common SNP greatly increased the risk for developing organ dysfunction when participants were fed a low-choline diet (OR = 2; P < 0.00005, based on 64 women studied) (20). We noted earlier that choline and folate metabolism are highly related...."
#49
Posted 24 December 2015 - 10:28 PM
Nutrigenomics and Cancer Prevention
http://www.ncbi.nlm....pubmed/24910810
"... Life is a series of intertwined biological signals. During the initiation and progression of cancer, several of these signals are modified to promote uncontrolled cellular growth. It is becoming increasingly apparent that some of the components in foods can have a marked influence on the risk of developing the initial and sustained changes in hallmark cancer signals. The ability of foods and associated constituents to influence the processes is linked to genetic variations that can influence the biological response in terms of the amounts reaching the molecular target(s) (ie, absorption, metabolism, and excretion) and also regulate the constitutive amount of the molecular target(s) requiring modification. Findings to date demonstrate that nutrigenomics and the downstream events (proteomics and metabolomics) and associated “-omics,” such as microbiomics, can have a significant impact on the relationship between dietary exposures and cancer risk/tumor behavior. However, the complexity of defining this interrelationship cannot be overemphasized because the thousands of food components and their common biological characteristics make it exceedingly difficult to unravel which constituent(s) is/are most critical for reducing cancer risk. The future likely will involve defining subgroups of individuals who benefit most from exaggerated intakes of selected foods or their component(s). As not all individuals will be expected to experience the same benefits, guidelines likely will need to be tailored based on selected genetic/epigenetic/transcriptomic variants. This approach also should be appropriate for those individuals who are at increased risk of cancer risk due to exaggerated intakes. Given the alarming increase in cancer worldwide along with other noncommunicable diseases, the impact of incorporating a personalized approach for using diet to curb risk holds enormous potential to improve quality of life, expand productivity, and reduce health care cost...."
#50
Posted 25 December 2015 - 06:14 PM
Interesting read. Independently by the correctness of the flow-chart this guy (a researcher in machine learning) provided by dissecting a patent application supposed to predict the most effective personalized nutrition plan (macronutrients) and exercise program for weight loss, I think this is a nice example of what is in principle possible by being guided by AI and prior to clinical assessment. Here we had humans doing the work, I believe we will be one day (sooner than later) be able to train a machine to do this for us for many diseases and personalized plans and likely much more effectively:
 personal diet and excercise.PNG 154.86KB
2 downloads
personal diet and excercise.PNG 154.86KB
2 downloads
http://rockstarresea...or-your-body-2/
The (dense) patent application is here:
http://www.faqs.org/...app/20100136561
#51
Posted 28 December 2015 - 05:32 PM
We all know cancer risk increases as we increase in particular consumption of meat (see e.g. 1, 2). Genetically I was looking at this here in relation to colorectal cancer.
But even more impressive is when you look at combinations of SNPs. Here the authors looked at some of the SNPs of the hugely important Cytochrome P450 enzymes which in particular “metabolize external substances, such as medications that are ingested, and internal substances, such as toxins that are formed within cells.”
Combinations of Cytochrome P450 Gene Polymorphisms Enhancing the Risk for Sporadic Colorectal Cancer Related to Red Meat Consumption
http://cebp.aacrjour.../7/1460.long#T4
What they find is very impressive: even if looking at 6 specific SNPs separately shows no particular risk effect, 3 variant combinations were found increasing risk with excessive red meat consumption up to almost 50x !!
“….In conclusion, we observed three combinations of CYP allelic variants that strongly predispose to CRC by interacting and enhancing the intrinsic predisposing potential of excessive red meat consumption. One of these combinations, ATGCGT, has a particularly strong effect on CRC risk …”
I am going to look at similar studies which the paper reports exist also on prostate cancer where I do carry an increased risk...
(1) http://www.ncbi.nlm....pubmed/15956652
(2) http://www.ncbi.nlm....pubmed/16507831
#52
Posted 29 December 2015 - 10:37 AM
I am looking at my magnesium status, where I seem having some irregularities probably related to absorption, and particular risks. The following study points to a particular gene mutation (Thr1482Ile polymorphism, rs8042919) and the risk of colorectal cancer:
The relation of magnesium and calcium intakes and a genetic polymorphism in the magnesium transporter to colorectal neoplasia risk.
http://www.ncbi.nlm....pubmed/17823441
“…We found that Thr1482Ile polymorphism significantly interacted with the Ca:Mg intake in relation to the risk of either adenomatous or hyperplastic polyps. Persons who carried ≥1 1482Ile allele were at greater risk of adenoma or hyperplastic polyps, particularly if they consumed diets with a high Ca:Mg intake. Among persons who carried ≥1 1482Ile allele, the inverse association with magnesium intake was further reduced, whereas high calcium intake tended to be related to a greater risk of adenoma…”
The study stresses the importance of a better understanding of nutrient-nutrient interactions when evaluating the interplay between personalized diet and SNPs.
“…We found that, as was consistent with our hypothesis, the intake of magnesium or calcium may be associated with a lower risk of adenoma only when the Ca:Mg intake is low and dietary vitamin D is high…”
I noticed a statement, particularly interesting to me in relation to magnesium absorption, which would support my strategy to supplement both with Vitamin D and magnesium together:
“…Therefore, absorption of magnesium may be significantly elevated when the vitamin D intake is high and the Ca:Mg intake is low. Because calcium is more sensitive to vitamin D than is magnesium, when both Ca:Mg intake and vitamin D intake are high, the absorption of magnesium is suppressed, but calcium absorption is substantially increased…”
Finally, this study also prompts investigation on how SNPs influence response to calcium which I will look after...
#53
Posted 01 January 2016 - 09:40 PM
Where do we find well structured information on the existing nutritional genomics public domain tools? It is a vast area but a must if you do serious nutritional research vs. my amateurish here ;-)
A nice compilation paper is:
Online Tools for Bioinformatics Analyses in Nutrition Sciences
http://advances.nutr...nt/3/5/654.full
"Abstract. Recent advances in “omics" research have resulted in the creation of large datasets that were generated by consortiums and centers, small datasets that were generated by individual investigators, and bioinformatics tools for mining these datasets. It is important for nutrition laboratories to take full advantage of the analysis tools to interrogate datasets for information relevant to genomics, epigenomics, transcriptomics, proteomics, and metabolomics. This review provides guidance regarding bioinformatics resources that are currently available in the public domain, with the intent to provide a starting point for investigators who want to take advantage of the opportunities provided by the bioinformatics field..."
Edited by albedo, 01 January 2016 - 09:42 PM.
#54
Posted 02 January 2016 - 11:13 AM
Though I do not carry the risk allele A on the FTO gene (rs9939609), hence I am not studying all this in detail, I thought you might be interested to these key studies on strong association to risk of obesity in child’s and adults and the importance of a Mediterranean style diet recommendation known to be beneficial from many other population studies. Note that the study (1) also mentions rs1121980 but do not report it due to the high linkage disequilibrium between the two. SNPedia reports that at least four other SPNs are also highly correlated to the two (rs9939973, rs7193144, rs9940128 and rs8050136)
- Fat mass- and obesity-associated genotype, dietary intakes and anthropometric measures in European adults: the Food4Me study
http://www.ncbi.nlm....pubmed/26620191
- A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity.
http://www.ncbi.nlm....pubmed/17434869
- A 3-year intervention with a Mediterranean diet modified the association between the rs9939609 gene variant in FTO and body weight changes.
http://www.ncbi.nlm....pubmed/19918250
#55
Posted 08 January 2016 - 04:06 PM
Since several years, I have a certain degree of dyslipidemia which I am trying to have under control in collaboration with my doctor but without using statins. First because of the secondary effects (even if statins are not the worst drugs you might take), second because a very atherosclerotic form of cholesterol (LP(a)), which I have relatively high, as well as the dangerous small size LDL particles, look unaffected by statins (1) and finally as I was pretty successful with natural interventions: diet, exercise and supplementation.
I try here to put together some findings and actions. Far to be complete and work in progress….
The first and most successful action has been lowering triglycerides (TG) by drastically cut on fast metabolized carbohydrates which basically went along with increasing (good) fats. Two VAP tests looking in particular at the size of LDL particles (small is bad, apoB level is also a measurement of small particles count) have shown a positive shift from a bad “pattern B” (smaller size) to good “pattern A” (larger size). It is well known (2, 3) that peak LDL diameters vary inversely with plasma TG’s levels. Should you not have a VAP test available, you might look also at the ratio TG/HDL where >3.8 is a strong indicator of the presence of small LDLs (4). While avoiding those fat cheeses in the evening, keeping my exercise schedule and supplementing with 1200-1800 mg of Red Yeast Rice (the only natural form of statin I allowed myself) I could to some extent lower my total-C, lower the total-C/HDL ratio, lower LDL, lower the apoB/apoA-I ratio and increase HDL. Also, when looking at my free fatty acid (FFA) profile, I noticed an increase in my monounsaturated/saturated FFAs ratio which is likely due in particular to the equivalent increase of the palmitoleic/palmitic FFAs ratio (also discussed in this thread). See also (5).
My dyslipidemia is classified as primary vs. familial, meaning I can primarily affect it by lifestyle and nutritional changes. In familial dyslipidemia, the bad pattern B phenotype has been associated to APOB and APOE polymorphisms. More recently the APO5 gene genotype has been found “…associated with increased plasma triglyceride and small, dense LDL predicts greater diet-induced reduction of LDL2, a haplotype-specific effect that is strongly correlated with both increased VLDL precursors and LDL4 products. Understanding of such diet-genotype interactions may help to elucidate mechanisms that are responsible for phenotype B and for its differential dietary responsiveness. This information may also ultimately help in identifying those individuals who are most likely to achieve cardiovascular risk benefit from specific dietary interventions ...” (6)
Unfortunately so far I have not been successful with the LP(a) (Lipoprotein(a)) which is particularly pathogenic and is currently considered to be one of the strongest genetic risk factor for coronary heart disease. My LP(a) remains stubbornly high around the 300-350 mg/dl (ref. <300). The LP(a) molecule is composed of an LDL cholesterol molecule and a glycoprotein, apolipoprotein(a). There are huge differences in plasma ranges between healthy individuals (even by a factor 1000) and it looks that pathogenity is possibly caused by the binding of oxidized phospholipids to the Lp(a). So I am trying to look at the genetics of LP(a) in view of a possible intervention (7, 8), e.g. looking at the LPA gene. Basically what I found is I do not carry the highest risk alleles in rs3798220 and rs10455872 (resp. C and G) but I am homozygous in the higher risk alleles in rs6919346, rs10755578 and rs9365171 (resp. C, G and C) which however are less risky for coronary disease (8). I do carry also other risky polymorphisms on the same gene. So genetically it is not a surprise I have a relatively high LP(a). Beside an expensive process called apheresis for which I will probably not qualify, high doses (1500-2000 mg) of niacin and low dose aspirin might be indicated (7,9) to lower LP(a). The problem I have with niacin is the effect (besides flushing) of strongly rising my homocysteine. I should maybe initiate a doctor supervised follow-up using high doses niacin and in parallel using high doses of l-methylfolate (e.g. 1000-5000 mcs) and other B vitamins to keep homocysteine at bay while looking at the hepatic enzymes as well (risky with niacin). I will check in my next consultation. I am already using a low dose aspirin from time to time.
I hope this helps.
- Statins Do Not Decrease Small, Dense Low-Density Lipoprotein http://www.ncbi.nlm....cles/PMC2929871 and HMG CoA reductase inhibitors lower LDL cholesterol without reducing Lp(a) levels http://www.ncbi.nlm..../pubmed/2530005
- An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing http://www.ncbi.nlm....pubmed/11588264
- Detection and quantitation of LDL subfractions http://journals.lww....actions_.5.aspx
- Managing Dyslipidemia: The Triglyceride/High-Density Lipoprotein Axis (Slides with Transcript, requires registration but you can try googling the title) http://www.medscape....warticle/582014
- Effects of dietary fatty acids and carbohydrates on the ratio of serum total to HDL cholesterol and on serum lipids and apolipoproteins: a meta-analysis of 60 controlled trials. http://www.ncbi.nlm....pubmed/12716665
- Dietary and genetic probes of atherogenic dyslipidemia. http://www.ncbi.nlm....pubmed/16166563
- Lipoprotein(a): resurrected by genetics http://www.ncbi.nlm....pubmed/22998429
- Genetic Variants Associated with Lp(a) Lipoprotein Level and Coronary Disease http://www.ncbi.nlm....pubmed/20032323
- Do We Know When and How to Lower Lipoprotein(a)? http://www.ncbi.nlm....pubmed/20842562
#56
Posted 10 January 2016 - 09:17 PM
A simple to read and quite recent (2014) review clarifying the terminology and giving also good references:
Nutrigenomics: Definitions and Advances of This New Science
http://www.hindawi.c...me/2014/202759/
Edited by albedo, 10 January 2016 - 09:18 PM.
#57
Posted 10 January 2016 - 09:19 PM
Nutrino and IBM Introduce Watson-Powered Nutrition Recommendations for Expectant Moms-to-Be
http://www.prweb.com...web13117207.htm
#58
Posted 16 January 2016 - 09:55 AM
Maybe not directly related to a personalized approach but the following article by Ben Best in the Dec. 2015 Life Extension Magazine, reporting on the 2015 American Society for Nutrition conference, is plenty of good information and references and include Ben's recommendations (he is a long time longevity enthusiast and expert and I used to go quite often to his web page in the past; it is no coincidence he has been hired by LEF). In particular his (good) high fat and low carb nutrition plan makes sense and indeed worked for me.
The 2015 American Society For Nutrition Conference
http://www.lifeexten...ference/page-01
Edited by albedo, 16 January 2016 - 09:57 AM.
#59
Posted 17 January 2016 - 10:22 AM
If we turn to be or think we are at increased risk of Alzheimer's disease (AD) copper status should be monitored, mostly if we have a copper metabolism irregularities. The following study is interesting and points to a preventive low copper nutrition plan. It also indicated the most important genes involved in the copper metabolism (e.g. ATP7B, ATOX1, and COMMD1) which we can look at.
Low-copper diet as a preventive strategy for Alzheimer's disease.
http://www.ncbi.nlm..../?term=24913894
"...The identification of individuals with a higher AD copper risk can be performed by 1 of 2 approaches. The first is a biochemical method based on the analysis of serum non-Cp copper, looking for values greater than the normal reference value of 1.6 mmol/L. The second approach is based on the sequence analysis of genes involved in copper metabolism (e.g., ATP7B, ATOX1, and COMMD1), identifying the loss-of-function variants that account for the genetic predisposition of copper dyshomeostasis. Unfortunately, current knowledge about copper dyshomeostasis in AD prevents us, at present, from developing a reliable strategy that could be used to identify individuals with a high risk of developing the AD copper phenotype. We are currently working in tandem with other researchers to bridge this gap..."
Note that the reference value in the abstract refers to the non-Cp (non-ceruloplasmin) value which is of course related to the serum value (e.g. see here)
This article from LEF (a bit old thought) can also be interesting:
The Copper Dilemma
http://www.lifeexten..._copper/page-01
Edited by albedo, 17 January 2016 - 10:31 AM.
#60
Posted 17 February 2016 - 10:28 PM
Closely related to personalized nutrition is the necessity to have easy to access and well curated databases which we the people/patients, doctors, nutritionists can access to link genetic information with specific diseases risks. I searched what exists and to date I found a number of sites which I wish to record here for the benefit of the discussion:
DIYGenomics (http://www.diygenomics.org/)
Genomera (http://genomera.com/)
Patients Like Me (https://www.patientslikeme.com/)
Personal Genome Project (http://personalgenomes.org/)
So far I tried only DYIGenomics which can easily link your personal genomic data e.g. via 23andMe, see: http://www.diygenomi...p/gen_data.php#: your data is loaded, compared to the reference and some literature list for the known SNP is given. I use it as a good resource for my research and I am sure much more will come up in future with higher resolution and in depth view.
Also tagged with one or more of these keywords: personalized nutrition, personalized medicine, nutrigenomics, nutrigenetics
Science & Health →
Supplements →
Regimens →
Personalized MedicineStarted by 9lives , 05 Nov 2021 |
|

|
||
Science & Health →
AgingResearch →
Biomarkers & Genes →
Attempting To Further Reduce Biological Age: hs-CRPStarted by Michael Lustgarten , 26 Sep 2021 |
|

|
||
Science & Health →
Brain Health →
Is anyone interested in building a nutrigenomics app?Started by Dominic F. , 16 Sep 2016 |
|

|
||
Science & Health →
Brain Health →
Supplement recommendations based on 23andme resultsStarted by ctmitchell , 08 Jan 2016 |
|

|
||
Science & Health →
Supplements →
Suggesting a Nutrigenomics section of the ForumStarted by world33 , 17 Nov 2015 |
|

|
0 user(s) are reading this topic
0 members, 0 guests, 0 anonymous users