.
Abstract
Ultraviolet (UV) irradiation from the sunlight is a major etiologic factor for premature skin aging. Long noncoding RNAs (lncRNAs) are involved in various biological processes, and their roles in UV irradiation-induced skin aging have recently been described. Previously, we found that the lncRNA RP11-670E13.6 was up-regulated and delayed cellular senescence in UVB-irradiated primary human dermal fibroblasts. Here, we performed further investigations of RP11-670E13.6 function. The results showed that this lncRNA directly bound to miR-663a and functioned as a sponge for miR-663a to modulate the derepression of Cdk4 and Cdk6, thereby delaying cellular senescence during UV irradiation-induced skin photoaging. Moreover, we found that RP11-670E13.6 may facilitate DNA damage repair by increasing ATM and γH2A.X levels. In addition, heterogeneous nuclear ribonucleoprotein H physically interacted with RP11-670E13.6 and blocked its expression. Collectively, our results suggested that the RP11-670E13.6/miR-663a/CDK4 and RP11-670E13.6/miR-663a/CDK6 axis, which may function as competitive endogenous RNA networks, played important roles in UVB-induced cellular senescence.
Introduction
The aging of human skin is caused by genetic and environmental factors. Among environmental factors, solar ultraviolet (UV) B (290–320 nm) and UVA irradiation (320–400 nm) are the main factors, causing atrophy of the skin, coarse wrinkles and leathery skin [1–3]. DNA photodamage and UV-generated reactive oxygen species (ROS) are the initial molecular events that lead to most of the typical histological and clinical manifestations of skin aging. [4–6]. Most DNA damage is repaired by functional repair systems in cells, once unrepairable and extensive DNA damage occurs, cells terminate proper division and enter a cell-senescent state [7]. Although numerous factors are involved in cellular senescence, the p53-p21 and p16CDKN2A (p16)–phosphorylated retinoblastoma protein pathways are best documented in maintaining cellular senescence and growth arrest [8].
Long noncoding RNAs (lncRNAs), which are more than 200 nucleotides in length, have been shown to play crucial regulatory roles in numerous biological processes [9, 10]. The mechanisms of action of lncRNAs are multifactorial and largely dependent on the specific intracellular localization of the molecule [11]. MicroRNAs (miRNAs) are a class of short noncoding RNAs (~22 nucleotides in length) [12, 13] that inhibit the expression of target genes by binding to the 3′ untranslated region (3′-UTR) of specific mRNA targets and hence degrade the mRNA or suppress translation [14]. In recent years, the “competitive endogenous RNA” (ceRNA) hypothesis has been proposed, and several studies have suggested the occurrence of interactions between lncRNAs and miRNAs [15–17], adding to the complexity of interactions between diverse RNA species. Despite rapidly rising interest in the expression and function of lncRNAs in cellular senescence [18–20], their potential implications in skin photoaging remain virtually unexplored.
In the previous study, we initially found that RP11-670E13.6 was up-regulated in UVB-irradiated HDFs and delayed cellular senescence through the p16-pRB pathway [21]. In this study, we further investigated the functions and the regulatory mechanisms of RP11-670E13.6 in HDFs. Our results provided important insights into the RP11-670E13.6/miR-663a/CDK4 and RP11-670E13.6/miR-663a/ CDK6 axis as ceRNA networks in UVB-induced cellular senescence. Moreover, we found that heterogeneous nuclear ribonucleoprotein H (hnRNPH) physically interacted with RP11-670E13.6 and blocked its expression.
Results
UVB up-regulated RP11-670E13.6 in a ROS-independent manner, and knockdown of RP11-670E13.6 promoted cellular senescence
RP11-670E13.6 is a lncRNA consisting of one exon of 348 bp and located upstream of the TRIM25 gene locus in chromosome 17 (Figure 1A). As shown in Figure 1B, RP11-670E13.6 expression was significantly elevated in UVB-irradiated HDFs over time and the greatest increase was at 24 h after UVB irradiation.
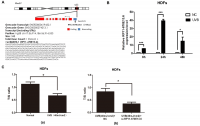
Figure 1. UVB up-regulated RP11-670E13.6 levels, and knockdown of RP11-670E13.6 promoted cellular senescence. (A) Schematic diagram of the localization of RP11-670E13.6. (B) Expression of RP11-670E13.6 in the UVB irradiation and control groups, as determined by qRT-PCR. Data are shown as the means ± standard errors of the means based on at least three independent experiments. (C) (a) UVB irradiation decreased the mean length of telomeres in HDFs at 24 h post-irradiation. (b) Knockdown of RP11-670E13.6 decreased the mean length of telomeres in HDFs at 24 h post-irradiation. Data are shown as the means ± standard errors of the means based on at least three independent experiments. P values were determined by Student’s t-tests. *P < 0.05; **P < 0.01; and ***P < 0.001.
In the previous study, we found that the ratio of senescent cells markedly increased following transfection with small-interfering RNA (siRNA) targeting RP11-670E13.6 compared with that of the negative controls (NC) [21]. It has been postulated that telomere shortening played an important role in photoaging [22]. Senescence in primary HDFs can be triggered by telomere erosion [23]. In this study, relative quantitative real-time polymerase chain reaction analysis confirmed the β-galactosidase staining findings, showing that the mean telomere length decreased in RP11-670E13.6 depleted HDFs at 24 h post-irradiation (Figure 1Cb). Moreover, the mean length of telomeres in UVB-irradiated HDFs decreased, suggesting that acute photodamage might contribute to early photoaging in human skin as a consequence of rapid telomere shortening (Figure 1Ca).
UV-induced ROS production is responsible for both clinical and biochemical manifestations of skin photoaging [24], and antioxidant enzymes, including catalase (CAT) and superoxide dismutase (SOD), are important for modulating ROS by scavenging free radicals in cells. To further investigate whether RP11-670E13.6 expression was required for modulating ROS generation or vice versa, we pretreated cells with a ROS scavenger (N-acetyl-Lcysteine, [NAC], 10 mM) before detection of RP11-670E13.6. As anticipated, 40 mJ/cm2 UVB exposure significantly increased ROS generation, and NAC caused a reduction in UVB-induced ROS generation (Supplementary Figure 1A). However, NAC had no significant effect on UVB-induced up-regulation of RP11-670E13.6 (Supplementary Figure 1B), neither generation of ROS nor SOD and CAT activity in UVB-irradiated HDFs were altered by RP11-670E13.6 reduction (Supplementary Figure 1C–1E).
Knockdown of RP11-670E13.6 induced DNA damage
To elucidate the molecular mechanisms through which RP11-670E13.6 affected UVB-damaged HDFs, we performed expression profiling of HDFs transfected with RP11-670E13.6 siRNA or siRNA NC using RNA-seq (Supplementary Figure 2A). Differentially expressed genes in RP11-670E13.6 knockdown HDFs were significantly associated with specific gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. In RP11-670E13.6-delepted HDFs, significantly enriched GO terms included biological processes, such as DNA replication (P < 4.9E-12), G1/S transition of the mitotic cell cycle (P < 2.1E-08; Figure 2A), nucleosome assembly (P < 1.3E-07), chromatin organization (P < 3.1E-07), and double-strand breaks (DSBs) repair via homologous recombination (P < 1.9E-06). Molecular functions, such as protein binding (P < 2.9E-06), helicase activity (P < 2.7E-05), and DNA binding (P < 3.9E-05) were also affected (Supplementary Figure 2B). Moreover, significantly enriched KEGG pathways included viral carcinogenesis (P < 3.7E-10), DNA replication (P < 3.8E-08), cell cycle (P < 6.6E-08), and transcriptional misregulation in cancer (P < 7.4E-07; Figure 2B). These findings were consistent with our previous study that knockdown of RP11-670E13.6 decreased HDFs proliferation and induced cell cycle arrest.
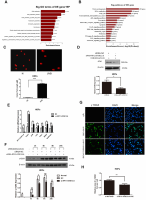
Figure 2. RP11-670E13.6 promoted DNA damage repair. (A) Top significant biological processes for genes whose transcript levels were increased in RP11-670E13.6-depleted HDFs. (B) Top significant Kyoto Encyclopedia of Genes and Genomes pathways for genes whose transcript levels were increased in RP11-670E13.6-depleted HDFs. (C) Comet tail length was quantified at 24 h after 40 mJ/cm2 UVB irradiation. Representative images are shown. Data are shown as the means ± standard errors of the means. (D) Representative image of western blotting results for the effects of RP11-670E13.6 on the expression of ATM protein in HDFs. (E) Relative expression of the indicated DNA damage-associated genes was determined by qRT-PCR in RP11-670E13.6-depleted HDFs and negative controls. Data are shown as the means ± standard errors of the means based on at least three independent experiments. (F) HDFs were mock treated or transfected with siRNA against RP11-670E13.6. Two days after transfection, the cells were UVB (40mJ/cm2) irradiated and analyzed for H2AX phosphorylation at the indicated time points by western blot. (G) HDFs were mock treated or transfected with siRNA against RP11-670E13.6. Two days after transfection, the cells were UVB (40mJ/cm2) irradiated and analyzed for H2AX phosphorylation at 24h post-irradiation by immunofluorescent staining. (H) Quantification of γH2A.X foci expressed as mean relative area per cell. Twenty nuclei from the HDFs transfected with RP11-670E13.6 siRNA and control siRNA were examined. P values were determined by Student’s t-tests. *P < 0.05; **P < 0.01; and ***P < 0.001.
Because the mRNA expressions of many genes involving in DNA replication and DSBs repair were significantly altered by RP11-670E13.6 depletion, we further examined whether RP11-670E13.6 played a role in the DNA damage response (DDR) in UVB irradiated HDFs. Comet assays revealed an increase in the tail length of HDFs at 24 h after 40 mJ/cm2 UVB exposure (Figure 2C), suggesting that the UVB dose of 40 mJ/cm2 could induce DNA DSBs in HDFs. Moreover, our results showed that RP11-670E13.6 depletion reduced the protein levels of ataxia telangiectasia mutated (ATM), which play a key role in UV damage signaling. (Figure 2D) [25, 26]. However, mRNA levels of ATM, in addition to many other genes involved in the DDR were significantly up-regulated by RP11-670E13.6 depletion (Figure 2E). It is well known that DSBs formation at late time points after UV treatment activates ATM kinase activity, which then contributes to the increase of phosphorylation of Ser139 of histone H2A.X molecules (γH2A.X) [27]. Our results showed that the phosphorylation of H2A.X was also decreased by treatment with an siRNA targeting RP11-670E13.6 in UVB-irradiated (40 J/m2) HDFs (Figure 2F). Immunofluorescence microscopic analyses showed that γH2A.X foci were also decreased in the RP11-670E13.6 depleted HDFs than in controls (Figure 2G). The relative area of γH2A.X was significantly lesser in the RP11-670E13.6-depleted HDFs at 24 h after UVB irradiation than in control HDFs (Figure 2H).
Cellular distribution of RP11-670E13.6 in HDFs
To further study the underlying mechanisms through which RP11-670E13.6 regulated cellular senescence, we examined the cellular distribution of RP11-670E13.6 in HDFs under physiological and UVB-irradiated conditions. In control cells (physiological conditions), fluorescence in situ hybridization (FISH) revealed RP11-670E13.6 in the nucleus, whereas it was detected in the cytoplasm after UVB irradiation (Figure 3A). By using cytoplasmic and nuclear RNA fractions from HDFs, we observed that RP11-670E13.6 is expressed in relative abundance in the cytoplasm after UVB irradiation, which confirmed the results of FISH (Figure 3B).
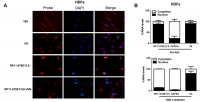
Figure 3. RP11-670E13.6 cellular localization. (A) FISH images showing localization of RP11-670E13.6 in HDFs treated with or without UVB irradiation for 24 h. (B) Percentage of nuclear and cytoplasmic RNA levels of RP11-670E13.6, U6 and GAPDH measured by qRT-PCR after subcellular fractionation in HDFs irradiated or not irradiated with UVB for 24 h. Data are shown as the means ± standard errors of the means based on at least three independent experiments. P values were determined by Student’s t-tests. *P < 0.05; **P < 0.01; and ***P < 0.001. FISH, fluorescence in situ hybridization; 18S, probe for 18S rRNA; U6, probe for U6 snRNA.
.../...
F O R T H E R E S T O F T H E S T U D Y , P L E A S E V I S I T T H E S O U R C E .
.













































