.
F U L L T E X T S O U R C E : bioXrv
Abstract
Genome-wide association studies often explore links between particular genes and phenotypes of interest. Known genetic variants, however, are responsible for only a small fraction of human lifespan variation evident from genetic twin studies. To account for the missing longevity variance, we hypothesized that the cumulative effect of deleterious variants may affect human longevity. Here, we report that the burden of rarest protein-truncating variants (PTVs) negatively impacts both human healthspan and lifespan in two large independent cohorts. Longer-living subjects have both fewer rarest PTVs and less damaging PTVs. In contrast, we show that the burden of frequent PTVs and rare non-PTVs is less deleterious, lacking association with longevity. The combined effect of rare PTVs is similar to that of known variants associated with longer lifespan and accounts for 1 − 2 years of lifespan variability. We further find that somatic accumulation of PTVs accounts for a minute fraction of mortality and morbidity acceleration and hence provides little support for its causal role in aging. Thus, damaging mutations, germline and somatic, can only contribute to aging as a result of higher-order effects including interactions of multiple forms of damage.
Introduction
Genome-wide association studies (GWAS) of human lifespan, including studies examining extreme longevity, parental survival, and healthspan, produced a number of gene variants associated with human longevity. For example, GWAS on centenarians consistently demonstrate the loci near LPA and APOE, FOXO3A, HLA-DQA1 and SH2B3 genes to be associated with longevity1. However, even in developed countries, centenarians currently represent less than 0.1% of the original cohort, and the genetic determinants responsible for the survival of general population remain poorly understood. Release of massive genotype and phenotype data by UK Biobank (UKB)2 inspired investigation of the relationship between genetics and longevity proxies, such as parental lifespan3 and healthspan within general population4. They confirmed most of the variants from centenarian studies and identified additional variants. However, the combined contribution of these variants could explain only a small part of lifespan heritability, at least as asserted from twin studies5. We hypothesized that the rest could be explained by the combined burden of rare damaging gene variants. Until very recently, only common variants could be probed in genetic studies due to sample size limitations. However, the large datasets such as gnomAD and UKB now allow assessing the effects of variants with minor allele frequency (MAF) lower than 0.1%.
These ultra-rare variants, most notably protein-truncating variants (PTVs), are known to be enriched for damaging alleles. They tend to have larger effect sizes and dramatically change gene expression and function. They are usually eliminated by purifying selection, but the small effective population size of the human population means that they are present in all human genomes. An inverse relationship between MAF and effect size was recently demonstrated for type II diabetes, an archetypal age-related disease6. Multi-tissue gene expression outliers were enriched with rare variants in the GTEx dataset7. Notably, PTVs represent a significant part of those variants. Most of the underexpressed outliers harbor rare variants and accordingly are subject to nonsense-mediated decay (NMD). Additionally, ExAC consortium demonstrated the nonsense variants with a high Combined Annotation Dependent Depletion score8, a widely used predictor of deleteriousness of single nucleotide variants, were enriched in singletons9. Although missense and non-coding variants may also be damaging, PTVs are substantially enriched for deleterious alleles. They also alter gene expression more dramatically than missense and untranslated region (UTR) variants10.
Increased rare PTV burden was shown previously to be associated with complex diseases, such as schizophrenia, epilepsy and autism11–13, whereas individual contributing genetic variants exhibited small effects. The burden of rare PTVs in genes intolerant to such variants (PI-PTVs) was tested for association across ExAC traits. For example, one study revealed a negative association with years of schooling (academic attainment) and a positive association with intellectual disability, autism, schizophrenia and bipolar disorder14. Notably, the age at enrollment was also negatively correlated with the burden of PI-PTVs referring to a possible association with lifespan. Overall, recent studies suggest that rare variants have a profound effect on complex traits and fitness. In this study, we focused specifically on the effect of germline PTV burden on longevity and disease-free survival, and estimated the effect of somatic PTV accumulation with aging on mortality and morbidity acceleration.
Results
Study design and data
We characterized the effect of ultra-rare damaging mutation burden on human traits associated with longevity. For the UK Brain Bank Network (UKBBN) postmortem samples, we run a survival analysis against the age at death15. For UKB subjects, we tested the effect of mutations on the remaining lifespan (or simply lifespan), that is survival within the available follow-up of eleven years. We also tested the effects of damaging gene variants on healthspan, that is the disease-free period (defined as the age when at least one of the following conditions is diagnosed for the first time: cancer, diabetes, myocardial infarction, congestive heart failure, chronic obstructive pulmonary disease, stroke, dementia, and death4). In addition, following the approach of16, we studied the effect of mutation load on parental survival (separately for the age at death for mothers and fathers), a useful longevity proxy in genetic studies.
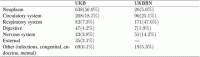
We selected a cohort of 40, 368 individuals from UKB with sequenced exomes who selfreported ‘White British’ and were of close genetic ancestry based on a principal component analysis of their genotypes2. Of those, 21, 742 (54%) were males with mean age 58.1 years (SD = 7.9, age range 40.2 − 70.6) and 18, 626 (46%) were females with mean age 57 years (SD = 7.8, age range 40.1 − 70.4) at the time of assessment. In this cohort, 1, 122 subjects died during the follow-up period of 11 years (2005 − 2016), mostly of cancer (Table 1 and Table S1). The UKBBN cohort included 1, 105 subjects of European origin excluding cases of suicides, accidents and cases of death with no abnormalities detected. Of those, 489 (44%) were females with mean age of 71.2 years (SD = 18, 16 − 103 years) and 616 (56%) were males with mean age of 67.7 years (SD = 17, age range 17 − 105 years). The cause of death was reported for 359 individuals. Most of the participants in this study were diagnosed with neurodegenerative diseases, e.g. Alzheimer’s, Parkinson’s, and Pick’s diseases.

As in14, we identified PTVs as variants that alter open reading frames of canonical transcripts, including splice donor/acceptors, stop codon gains and frameshifts. To address the relation between the frequency of mutations and their effects on lifespan, we binned gene variants from whole-exome sequencing (WES) datasets according to their frequency: 1) MAF < 10−4; 2) 10−4 < MAF < 10−3; 3) 10−3 < MAF < 0.01; 4) 0.01 < MAF < 0.2. We defined PTV exome burden as the total number of PTVs within the MAF bins (Figure S1 for PTV burden distribution in the MAF bins).

Figure S1:
Histograms of PTV burden distributions depending on the PTV frequency.
Survival analysis
We examined the association of PTV load in the defined MAF bins against the selected longevity traits (i.e. survival in UKB and UKBNN, and healthspan and mother’s and father’s age at death in UKB) using Cox proportional hazards (PH) models. We used sex and genetic principal components as covariates to account for the effects related to the population heterogeneity. We additionally used age as a covariate for the UKB lifespan during the follow-up period.
The mortality and morbidity risk models returned the Cox regression parameters that were consistent with well-established mortality patterns. For example, the UKB survival model produced the regression coefficient Γ = 0.093 per year for the age of first assessment, very close to the mortality and morbidity acceleration rate (of approximately 0.09 per year in UKB cohort4) and hence consistent with the Gompertz mortality law. The Cox regression coefficient for sex was 0.47 for males in UKB and 0.29 in UKBBN. Under the constant mortality acceleration, this would correspond to approximately 3 − 5 years difference in life expectancy. Women in the UK (the population relevant to this study) live longer than males, although the gap between the sexes has decreased over time and is now 3.7 years17.
.../...












































