Scientists have developed a new open source transcriptomic aging clock and published their work as a pre-print [1]. The newly identified rejuvenating drugs and gene perturbations could be applied in regenerative medicine and longevity therapies, and the aging perturbations could find use for anti-cancer therapies.
Age-shift learning—one model for every platform
Pasta (Predicting age-shift from transcriptomic analyses) was trained on 17,212 healthy profiles from 21 bulk-RNA-seq or microarray studies. A ridge-regularized classifier learned from rank-difference vectors of same-tissue sample pairs ≥40 years apart, capturing the ordering of age-responsive genes rather than their absolute counts. In leave-one-dataset-out (LODO) tests this 40-year age-shift model outperformed the regression baseline and surpassed the previous multi-tissue benchmark, MultiTIMER [2], in 6 / 8 LODO datasets and 5 / 6 independent datasets.

When single-cell data were aggregated into pseudobulks, Pasta retained the highest Pearson correlations, illustrating true cross-platform portability. Because ranks are platform-agnostic, the same coefficients apply, without re-alignment or batch-correction, to raw or normalized bulk, scRNA-seq and L1000 count matrices.
p53-anchored biology linked to senescence, stemness and cancer
Gene-set enrichment placed p53-mediated DNA-damage response at the core of the model: positive weights were dominated by CDKN2A/p16, ZMAT3 and other p53 targets, while miR-29 targets with anti-p53 activity carried the strongest negative weights. Functionally, Pasta separated senescent from proliferative or quiescent cells in 24 / 30 RNA-seq datasets and tracked a 450-day fibroblast passage toward replicative arrest almost linearly (PCC ≈ 0.90). At the opposite extreme it assigned exceptionally “young” scores to embryonic stem cells and to fibroblasts during OSKM reprogramming, while scores rose during directed hepatocyte differentiation, capturing both directions of the stemness-senescence continuum.

Applied to TCGA cancer patient samples, Pasta’s age scores correlated more strongly than chronological age with histological tumor grade and stratified overall survival in adrenal, sarcoma, thymoma and other cancers, underscoring clinical relevance.
Chemical and genetic modulators of cellular age
Screening ~3 million CMAP L1000 profiles [3], the authors flagged 259 Aging and 59 Rejuvenating compounds. Pro-aging hits were highly enriched in FDA-approved chemotherapeutics (e.g., mitoxantrone, gemcitabine, doxorubicin), whereas rejuvenating compounds contained HDAC, MEK and GSK-3 inhibitors, compound classes often used in chemical reprogramming cocktails.
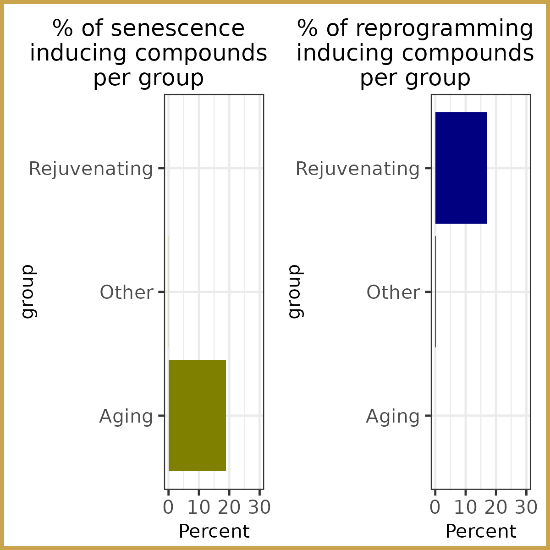
Two predictions were taken to the bench: the antifolate pralatrexate (4th Aging hit) triggered SA-β-gal staining, p21 induction and irreversible arrest in A375 melanoma cells, while the natural product piperlongumine (20th Rejuvenating hit) up-regulated OCT4, SOX2 and NANOG in PC3 prostate cells, confirming a stemness-promoting, age-reversing profile. Extending the same pipeline to CRISPR, shRNA and over-expression constructs uncovered 841 Aging and 54 Rejuvenating gene perturbations. Aging drivers included CCNA2 knockout and KRAS/BRAF over-expression, mirroring cell-cycle arrest and oncogene-induced senescence; while youthful scores included MYC over-expression and TP53 loss, echoing known reprogramming and tumour-suppressor pathways.
Molecular traits that prime cells to respond to aging or rejuvenating cues
To pinpoint intrinsic features that dictate how a cell responds to age-regulatory cues, the authors computed aging- and rejuvenation-propensity scores, which corresponds to the average response of a given cell line to all identified age-modulatory chemical and genetic perturbations. They then correlated these scores to omics measurements for 19 cell lines in DepMap. Mutation analysis singled out TP53: lines bearing two inactive alleles almost never aged, those with one allele aged moderately, and wild-type lines aged readily (PCC = 0.88). Aging-prone lines were wired for oxidative stress: their proteomes were enriched for mitochondrial-translation and respiratory-chain factors, their transcriptomes echoed the same signature, and their genomes often carried extra copies of TRAF6-IRF7 and IFNA loci—configurations that amplify ROS and interferon-driven SASP signalling. Rejuvenation-prone lines, by contrast, over-expressed spliceosome, histone-acetyltransferase and other chromatin-remodelling proteins and frequently lost PRC2 and HDAC copies, genetic changes known to lower epigenetic barriers to induced pluripotency. These results may help design more personalized and effective rejuvenation and anti-cancer therapies.
Literature
[1] Salignon, J. et al. Pasta, an age-shift transcriptomic clock, maps the chemical and genetic determinants of aging and rejuvenation. 2025.06.04.657785 Preprint at https://doi.org/10.1101/2025.06.04.657785 (2025).
[2] Jung, S., Arcos Hodar, J. & del Sol, A. Measuring biological age using a functionally interpretable multi-tissue RNA clock. Aging Cell 22, e13799 (2023).
[2] Subramanian, A. et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 171, 1437-1452.e17 (2017).








































