Wow, that is good, you seem having wrote this for me as I for one was starting to get lost and this post helped :-) I start to get what you are heading to.
It maybe be trivial but what evidence do we have that a non-OSKM strategy does the same?
I think there are going to be many spinoffs from this such as regenerative medicine. Shooting for longevity is going to provide research into many areas we didn't expect.
I think this topic is a bit thick for most of us, at least me, so your input is valuable and telling about what needs to be covered. You also ask a very important question, "what evidence do we have that a non-OSKM strategy does the same?"
You're asking the right questions, and you also in a roundabout way answered the same. "...Importantly, we also demonstrate, by using this platform, the induction of cancer stemness properties in MCF7 cells..." Plus as we covered, we don't have a drug-inducible, polycistronic expression cassette that permits rapid and potent induction of OKSM of the individual factors. This approach is a nonstarter for humans, and there are cancer risks. To answer your question head-on, yes and no.
This study shows the context of the environment can induce cell plasticity and there are specific epigenetic locations being activated. I believe advances in regenerative medicine and spinoff applications to aging will emerge from this study. I never imagined a mature cell could regress and play another role. So, this is yes but not applicable to an entire organism at least where this study left off.
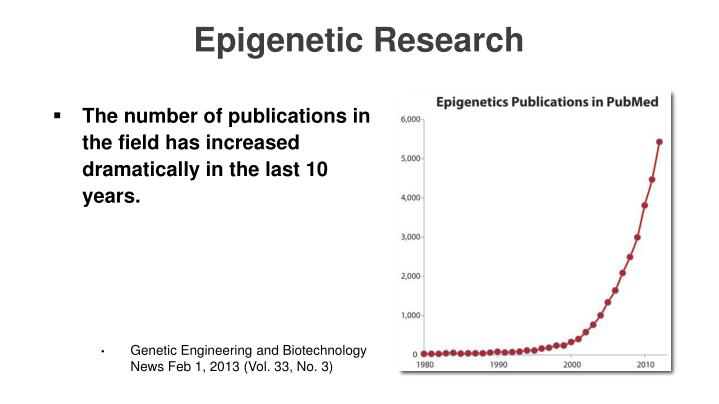
Other epigenetic reprograming avenues are discussed in "3.2. iPSC reprogramming vs other longevity/rejuvenation technologies" Now respectfully consider this, and I mean no disrespect because that author covered a lot of relevant topics, but the insights given here are somewhat dated. Submitted: October 27th, 2015.
Hard to believe 3 years is a little behind the curve, but by comparison 15-years ago this topic was moving at a relative snail's pace. Let me give you a sense of how quickly this field is emerging and this chart is in no way up to date.
Your observation of tumors and cancers from the OSKM technique is not an entirely foregone conclusion but a topic of considerable concern. It appears if a cell is overexposed to OSKM factors, prematurely terminated from exposure or is predisposed to such cancer states, (or should I say already on the fence,) increasing that cells plasticity could have an undesirable effect. So this technique appears to have the characteristics of a balancing act. The flip side, it also shows the possibility of correcting some cancer states. The OSKM technique has opened so many doors.
"Indeed, our previous study demonstrated that premature termination of in vivo reprogramming leads to kidney cancer development through altered epigenetic regulation. Consistent with the partial reprogramming state, these cancer cells lose kidney cell-specific molecular signatures while they partially acquire the trait of embryonic stem cells (ESCs) including self-renewing capacity. Notably, these cancers resemble Wilms’ tumor, which is the most common childhood kidney cancer. Furthermore, these cancer cells were readily reprogrammable into iPSCs that are capable of differentiating into non-cancerous kidney cells. These results raised the possibility that reprogramming-associated epigenetic regulation has a significant impact on childhood cancer development, which is also in agreement with recent observations that childhood cancers harbor relatively few genetic mutations. However, the functional significance of epigenetic regulation related to cellular reprogramming remains largely unclear in adult cancer development."
I see a few bright spots in the field that I think will help take on a guiding role.
"By splicing animals together, scientists have shown that young blood rejuvenates old tissues. Now, they are testing whether it works for humans."
After millions of dollars were dumped into this idea Conboy revised the experiment so the blood could be turned off. They would allow enough time for a 50/50 blood exchange and disconnect the mice. This eliminated the influence of shared organs which was thought to be skewing the data. As expected, the old mouse blood had a devastating effect on the young mice and on the flip side old mice hardly benefited at all. Their research team has begun to investigate what factors in the old blood that might cause the young mice to decline. I think their insights will prove to be very illuminating within the epigenetic reprogramming topic of this forum.
So what this new experiment appeared to indicate is the old mice were benefiting from the young mouse's organs, i.e., lungs, liver, kidneys and so on. It was also thought the old mice benefited from a dilution of the old-mouse inhibitors that adversely affected the young mice. So this is where I think this comes full circle back into this forums epigenetic topic, understanding the undesirable effects is as valuable as the positive ones and mapping those epigenetic markers needs to commence.
I think this is precious research.
So since these mice are bred to be genetically identical to avoid tissue rejection. My belief is the comparative epigenetic profiles of the joined, and intermittently connected mice will prove very insightful into which epigenetic locations are of interest to study. Certainly which epigenetic positions you might want to inhabit that caused tissue and organ damage to the young mice.
I don't think its necessary to take on a full OSKM factor approach but it does ring many of the right bells. The trick will be is how to regress the cell without losing tissue identity or leave a cell teetering between cell states. If a cell already has a tendency to move towards a cancerous state and many cancers are now being found to resemble earlier states of cell progression, this might be a huge hurdle. How can you know if the patient might be predisposed to particular cancer?
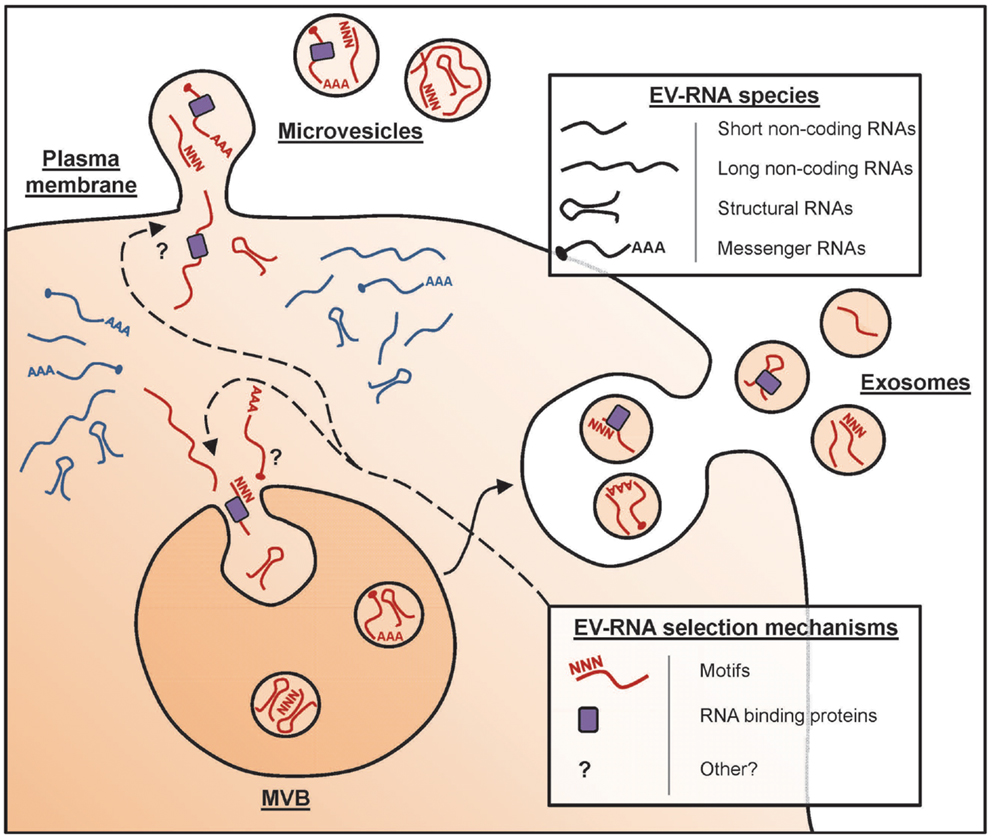
So I lean more towards a soft selective approach whereas the full OSKM factor approach opens too many doors of chance. I believe that in the UC Berkeley Conboy study not only were the old mice benefiting from the young mouse organs but also from constant exposure to their young extracellular vesicles and hormones helping to improve tissue regeneration and plasticity. See "
Extracellular vesicles and aging"
So along the murine parabiosis model here are two studies recently published.
So since I'm not a scholar in epigenetics like many of you, these studies take awhile to digest and hence an investment of time is required. Also once you think you have a firm grasp the next paper appears and changes the landscape. So, on the one hand, I experience some frustration, on the other as an optimist I see the rapid publication of papers to be a great thing even if I have to disregard some ideas from a previous article.
As always JMHO
Bryan